Be the best in bisulfite analysis
Terminate the controversy in mRNA m5C
I have been working with mRNA m5C for over four years, unbelievable. We began our research at the end of 2016, when the workflow of RNA m5C was not optimized. As other competitors, we were curious about the existence of mRNA m5C.
There were some challenges for us at that time:
-
firstly, the commercial kit is optimized for tRNA and rRNA BS-seq, but not for mRNA. With the standard protocol, we often have a conversion rate between 95% to 98%. Such a condition is fine for known sites in tRNA and rRNA, however, it’s disaster for us while applying it on mRNA. Based on binomial distribution, when the number of cytosines increase, we much more chance to get artifacts.
-
the bioinformatic tools available far from satisifying us. They are not fast enough, noisy, and heavy. Although most of the software are open-source, we are hard to modify their codes without fully understanding the logics behind them.
So our decision was to optimize the protocol (by Dr. Tao Huang) and to try to set up a new pipeline (by me). I guess Prof. Zhang did not expect such a huge project at that moment. In fact, we firstly began with meRanTK, but I gave up after several days. It so lucky that we had the tools for setting up one - we had the Pysam API of htslib (or Samtools) to handle the reads. That’s enough for us to translate our logics into codes. In the the Spring Festival of 2017 (yes, I love the long holiday), I gradually finished the prototype of the pipeline. To accerlate the computation, I introduced multiprocessing into the pipeline, for which I suffered a lot for the synchronization issue.

But we were stuned no long after we prepared to send our manuscript. Just after the holiday, the first mRNA BS-seq profile paper published on Genome Biology (Amort et al., 2017), pointing out that there can be thousands of m5C sites on mRNA in mammalian cells. No long after the first publication, another paper published on May reported that in HeLa cells and mouse tissues, thousands of mRNA m5C sites are contributed by NSUN2. Things got more interesting in July. The third paper published on Genome Res. from Frank Lyko (he set up the first transcriptome wide BS-seq protocol in 2008), argued that the mRNA m5C sites found in mammalian transcriptome were not real and just came from the artifacts caused by insufficient conversion, beacuse they didn’t find a convincible stoichiometric measurement for mRNA m5C.
We were scooped, but nowadays, I thanks for the scoop. We have to make a break or our works will be meanless. It forced us to turn to optimize our pipeline. In fact, in Lyko’s paper, they used some filter assembled what I used in my final pipeline (something like C-cutoff, but not systematically). The had beautiful experiments. If you zoom in the motif in the main figure, you can indeed found a 3’ G-rich motif (but they ignored). The major problem is, they missed in data analysis: they made mistakes in dropping the multiple alignments, which resulted in a great loss in m5C sites. And this is my conclusion based on my analysis of the differential sites between our pipeline and theirs. During the optimzation, I introduced other features, like Gini-index and signal-noise ratio, which are very helpful in the analysis. In our paper accepted by NSMB, we sucessfully clearified that NSUN2 is indeed a mRNA m5C writer for tRNA-like sequences. And thanks Alexandra Lusser for the comment on the same issue.
Set up a new paradigm in NSUN6 study
It was just the beginning of our mRNA m5C journey. In our NSMB article, it is obvious that except for NSUN2, there should be another enzyme contribute to the methylation (also mentioned in the comment). This enzyme should favor a 5’m5CUCCA-3’ motif. And now we know that this enzyme is NSUN6, another NSUN protein responsible for tRNA 3’ end methylation. It’s another story that three papers about that crahsed within two months for that…
But, don’t you think it’s boring? If the story is: we found a new enzyme; we had some barplots; we drew some lines, some boxplots, and some scatters; we had some Western Blots; and so on.
It’s fortunate that we have a the crystallization of NSUN6-tRNA complex! In Nov. 2018, Kappel, a Ph.D. student (now a Post-doc in Broad institute) from Das Lab published a RNA-protein complex simulation method (RBP-denovo) based on Rosetta. This method enable us to predict the folding status of RNAs dock to a protein. Personally, I love Foldit, and I am a long-term computational contributor to Rosetta@home. It’s a great chance for me to try so!
After a short struggle, I successfully set up Rosetta and figured out how I should do (in a mid-night, 5:00 am). Then I integrated a lot of protein simulation (comparative modelling), and RBP folding in the study. I found two possible models for NSUN6-mRNA complex: (1) mRNA might interacts with NSUN6 in a tRNA-like shape, or (2) mRNA laid tightly on the surface of PUA domain. To clearify the model, I schemed some mutagensis experiments to verify that, and I finally made sure that a tRNA-like process should be more reasonable. I also tried to use the predicted structure to guide us to design a NSUN6 variant that can distinguish mRNA and tRNA.
I guess this should be a new paradigm for us in RNA modification study: with simulation, we can focus on some residues of the RBPs, and we can design proteins to help us understand the biological processes. No long after my trials, AlphaFold published its achievements.
Three years!!!
In fact, we initiated our developmental mRNA m5C study half a year before the NSUN6 study. I don’t remember why Prof. Zhang decided to profile m5C in embryos at that moment. But we underestimated how long we will spent on it.
We won the bet. There are indeed a great enrichment of mRNA m5C in the embryos. It should be surprising, We guessed we had the chance to be accepted by the top journals. Yes, we did, closely.
We firstly submitted it to Nature, on the January of 2019. Our article was successfully sent to the reviewers, however, none of them seemed to have a practices in mRNA m5C. After the rejection, we sent the manuscript to Science. We entered the revision, but was just the beginning. One month latter, we received a 17-pages long comments. One of the reviewer, revised everything, even a comma, in our manuscript (I admired to him/her so much and this reviewer made a great impact on my working style). The major concern from the reviewers was that some of our conclusions were not purdent enough. We took a long time to handle these issues. However, we were scooped agian. Yang et al. published a similar paper on Mol. Cell a head of our re-submission. We were rejected. We then tried NSMB but the reivewers didn’t support our publication.
We decided to append some data (from human) and modify the paper to be a purely resource paper. We merged the evolutionary analysis we planned to publish independently in the new version. But this time, our manuscript failed to enter a revision sections. We tried many many journals. We were exhausted and sent it to Nat. Commun. finally on September, 2021, over 2.5 years after we had the first submission. Luckly, two of the reviewers gave us positive comments, but one of them seems unpleasant. He/she had only one comment that we should provide the stoichiometric reulst for the data, although that was totally not necessary. We carefully modify our manuscript and resubmitted to the editor. After >50 days’ suffering, we eventually received editors’ email telling us the manuscript was in principle accepted. I feel numb, rather than happy. Now we were liberated.
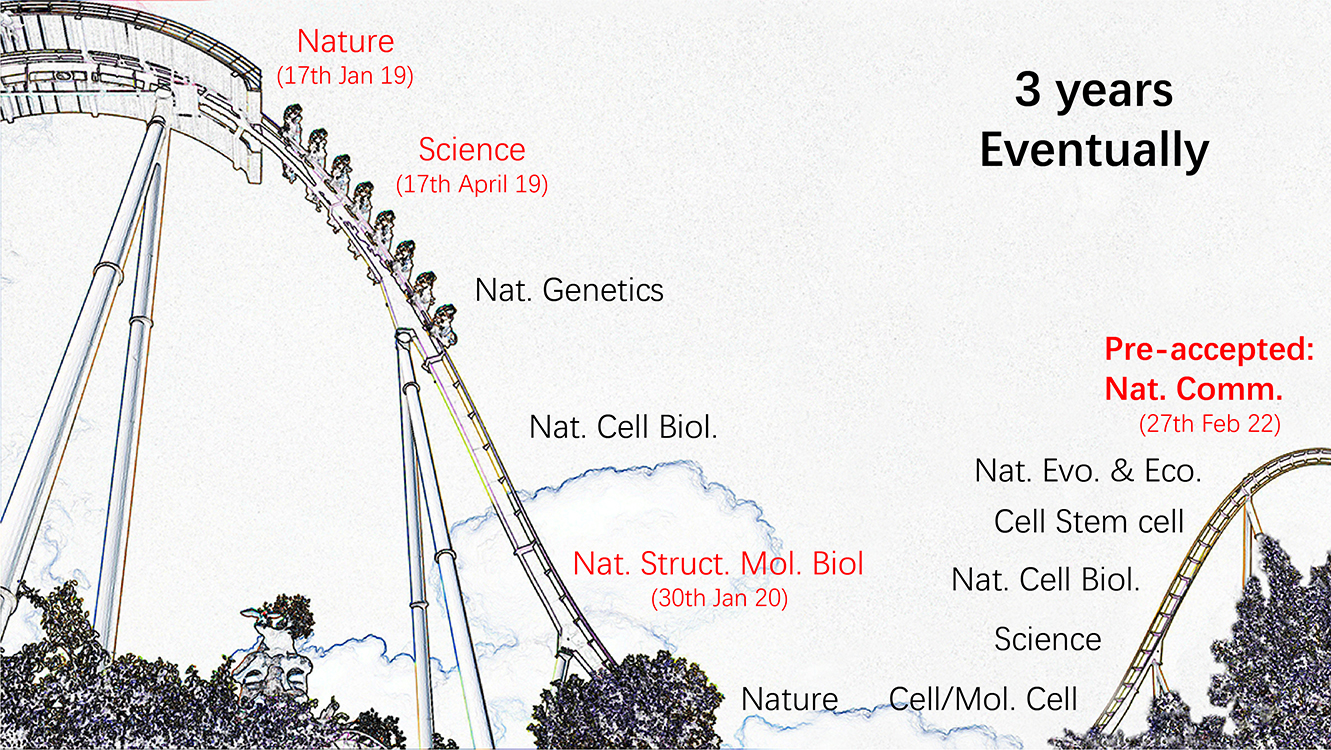

The hard road of mRNA m5C study
It’s 3 years past after we published our first m5C paper. Our pipeline stands in third-part tests and is graduately acknowledged by other labs as a gloden standard.
If I have to estimate the progress in these field these years, I might answer that, probably we just jammed. It’s still hard for us to learn the the real biological role of m5C and other modifications. We don’t have suitable techniques to exclude unwanted interferences in our experiments and analysis. For example, we often don’t have consisted methylation changes when somebody claims that m5C might involve in mRNA stablility control; although we have data showing that m5C can indeed have higher affinity with ribosomes, we don’t biophysic data to support our conclusions; we are unable to split the functions of mRNA m5C and tRNA m5C; we have no idea to figure out the function of single site modifications; … We don’t agree to some of the conclusions by other groups, but that’s not welcomed to have battles in this field, unlike that in evolution.
This field changed in these years. The editors and reviewers ask for functions more and more frequently. However, we don’t have enough data to support a clear “function” for most of the cases. We are strict with our conclusion, but we are unable to let others be prudent. The field, mRNA m5C, is too young for those requirements.
Sadly, our m5C group (me, Dr. Tao Huang, and Wanying Chen) are going to leave. We have tons of data, but we have no time to completely tide and publish. We are waiting for our followers to finish them. Maybe I will be back, when I have new paradigm for it.
Enjoy Reading This Article?
Here are some more articles you might like to read next: