The NSUN6 poject
Traditional molecular biolog plus simulations
In breif
- We verified that NSUN6 is a key contributor to mRNA m5C.
- NSUN6 favors a 5’-m5CUCCA-3’ motif and usually methylates cytosines in a small loop adjacent to a longer stem.
- NSUN6 has different activity in different types of tissues/cells.
- NSUN6 methylates mRNA in the cytoplasm, while NSUN2 methylates mRNA in both nucleus and cytoplasm.
- We profile the m5C in polysome fractions, and we found that m5C enriched in fractions with fewer ribosomes.
- We use simulations (RBP-denovo) to study the interactions between NSUN6 and mRNA.
- Based on the structure model, we try to mutate some residues to investigate the feature of the NSUN6-mRNA complex.
- We use high-throughput
mutation asssayto study the substrate recognition of NSUN6. - We design a new
tRNA BS-seq pipelineto study the changes of m5C levels on tRNA and mRNA.
About RBP structure simulation
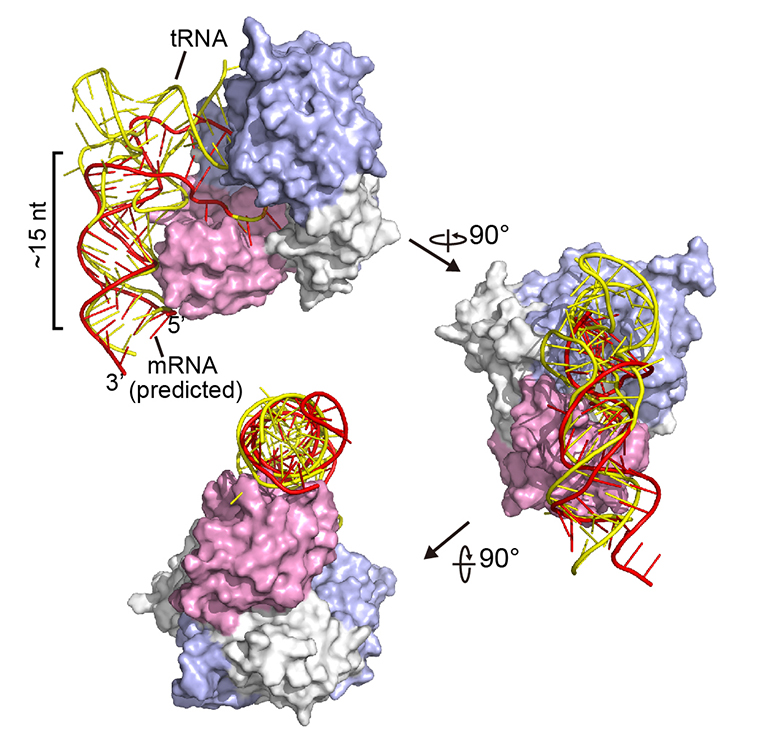
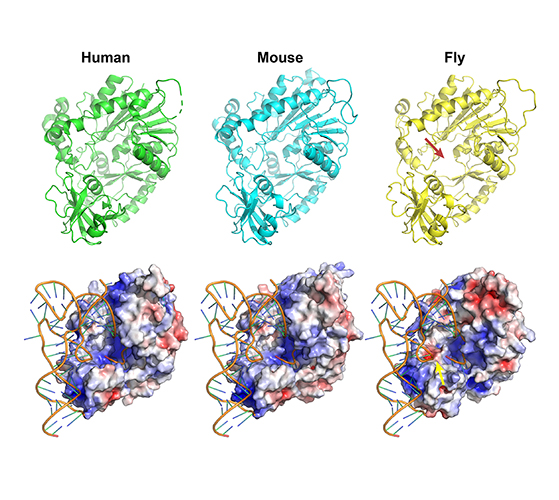
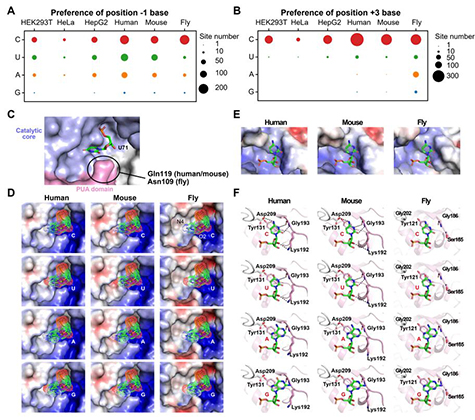
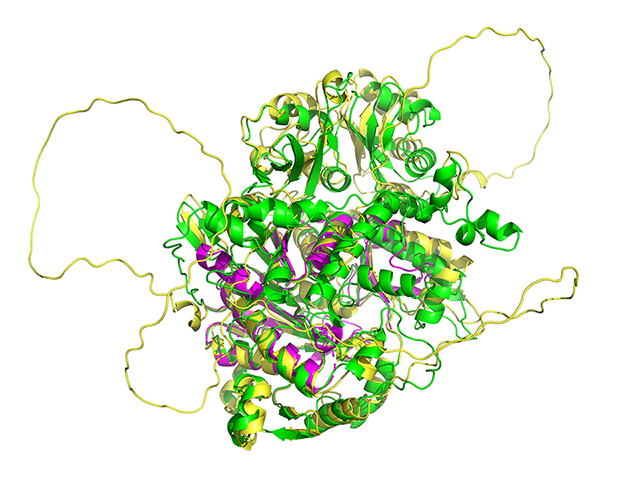
For a long time, I have a concern about our technique of simulation in biological studies: we can use simulation to boost our design in experiments, but how can we use simulation to directly understand our nature? An obvious thing is, if the object is hard to be finished/validated via experiemnts, so how can we know we are doing the correct things with our algroithms running in our computers? If the object is simple in reality, what is the meaning of simulation itself?
In 2020, the 14 th Critical Assessment of protein Structure Prediction (CASP), AlphaFold exhibited its fantastic performances in the test. It seems that if the RMS of the predition were high enough (>90%), the preditions can be treated as native structures. I was once stimulated, but quickly depressed by the structures of some big, unknown domains.
Back to this poject. This project is launched much early than CASP 14th in 2019, when RNP-denovo was just published one year ago. And luckly, we have the structure of the NSUN6-mRNA complex. It’s not an accidence for me to combine these two things. I have once be a player of Foldit and I am a long-term compuational contributor to Rosetta@home. I have been thinking about how to use the amazing utilities in my research. Here is the chance.
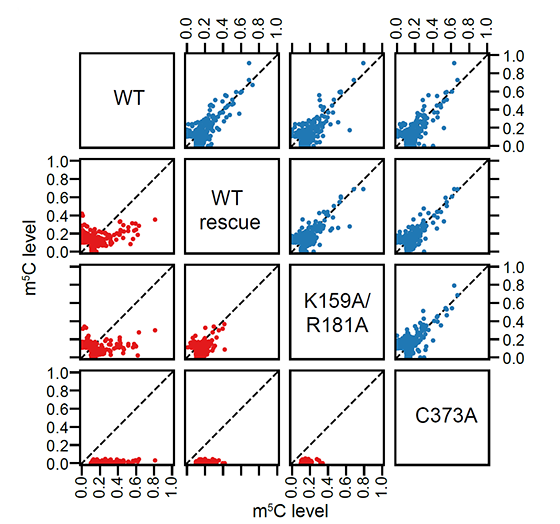
Now, I would like to set up a paradigm: if we have a structure model of RNA binding protein structure and if we have a reference structure of the RNA sequence (not so necessary), we can simulate the complex of RBP-RNA complex, and this will help us in learning the process and preference of RBP-substrate recognition. More importantly, we can be inspired to modify some residues to make the RBP different. In this project, what I want to do is to create a NSUN6 mutant that can only methylate mRNA or tRNA. This purpose comes from the challenge in RNA modification studies, we are unable to distinguish the functions of a modification writer in three aspects: RNA binding, RNA modification itself (a sub-issue is the different types of RNA), and others. We are annoyed that the editors and reviewers always ask for the function of the modifications, but to be purdent, it’s too hard for us to accurately locate the function without any interference, why others can pubish casualy? We finally generated a NSUN6 variant that can methylate mRNAs normally, but have a weaker methylation activity on tRNAs. Standard structural analysis also performed to learn the preference of NSUN6.
However, this paradigm is still limited in practices:
-
High-quality protein models, in complex or in apo state, are available in most of the cases. And for some proteins, the apo state is different from that with a substrate.
-
We tried to build up the simulated models for protein with missing reference structure. However, the current simulation tools, include
RosettaandAlphaFold, are unable to draw structures without any prior knowledge. For example, the C-termial of human NSUN2 protein has no homology structure available in our database, and hence bothRosettaandAlphaFoldfailed to simulate that part of the protein. -
We still don’t have a complete theroy to guide us how to adjust parameters, and how to select the right model for further simulation. And bugs in the softwares…