mRNA BS-seq pipeline and NSUN2
We still don't know the function and mechanism behind it
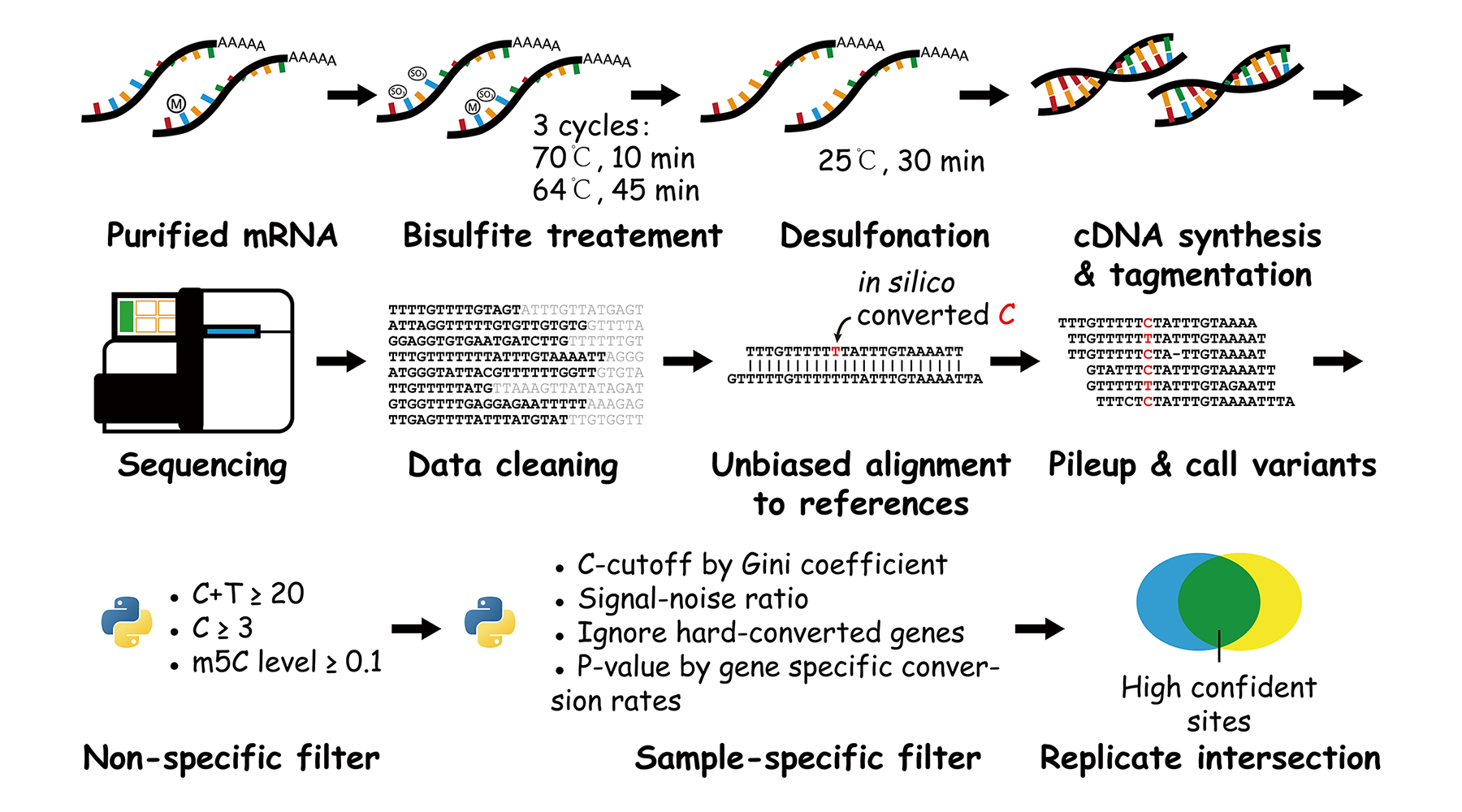
The schema of our optimized workflow.
In breif
- We optimized the experimental protocol to improve the conversion rates in mRNA BS-seq.
- We analyzed the pattern of artifacts in BS-seq and located the source of false positives.
- We introduced
Gini-indexandsignal-nosie ratiointo analysis to estimate the status of library conversion. - We introduced
C-cutoffandgene-specific conversion ratesinto our filter to remove false positives. - We set up a
new pipelinewith high flexibility, robustness and parallelization level for mRNA BS-seq. - We validated that NSUN2 is one of the major m5C methyltransferase acting on mRNA.
- We profile mRNA landscapes in multiple human/mouse tissues.
- mRNA m5C might relate to translation control.
Where are the noise?
It’s clear now that the noise in BS-seq come from many places:
- The human-introduced Cs in hexamer.
- The regions hard to be converted, especially some highly structured rRNA regions.
- The false alignments and bases with bad sequencing qualities.
Solutions
The pipeline I designed have considered most of the situations:
- Hexamer can be removed by trimming the bases at the end of reads.
- We should use
gene-specificconversion rates rather than aglobal conversion ratemeasured by spike-ins or the whole libray. Only analyze annotated regions. - Estimate the conversion status by
Gini-indexandsignal-nosie ratio, and determine theC-cutoffthat remove reads have more than a certain number of reads. - Optimize the alignment step to reduce false alignments. Normally, that should not be a matter.
- SNPs can only destory m5C, but cannot create m5C in BS-seq.
- Statistical tests have little effect on our analysis if we use a filter with high coverage and level.
In my hands, I guess we can let the sites with 5% methylation go, but we normally use a 10% cutoff for the biological importance of a site. BS-seq is very sensitive and reliable if you work with it correctly.
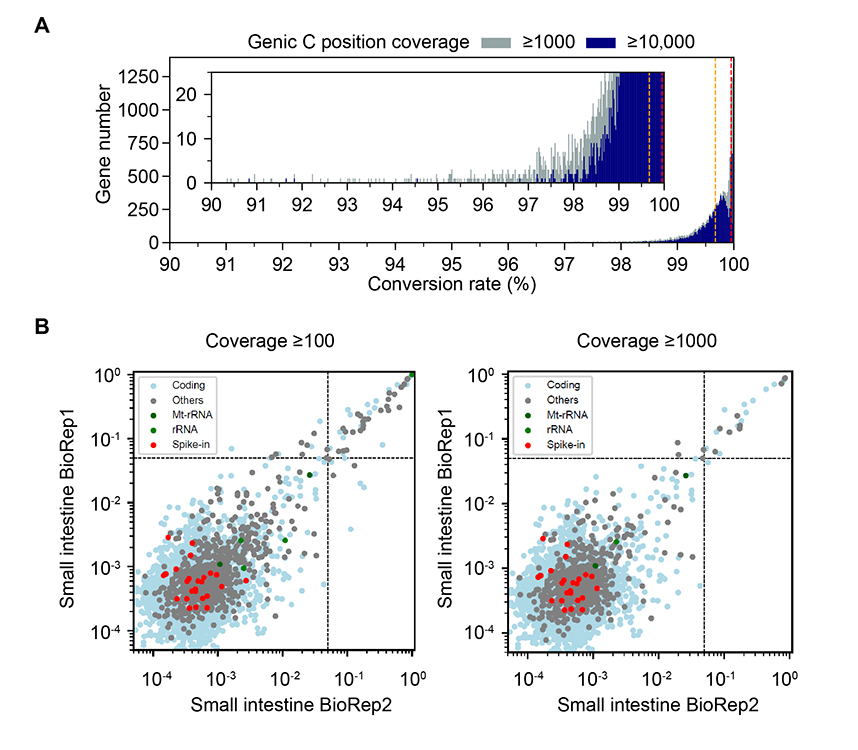
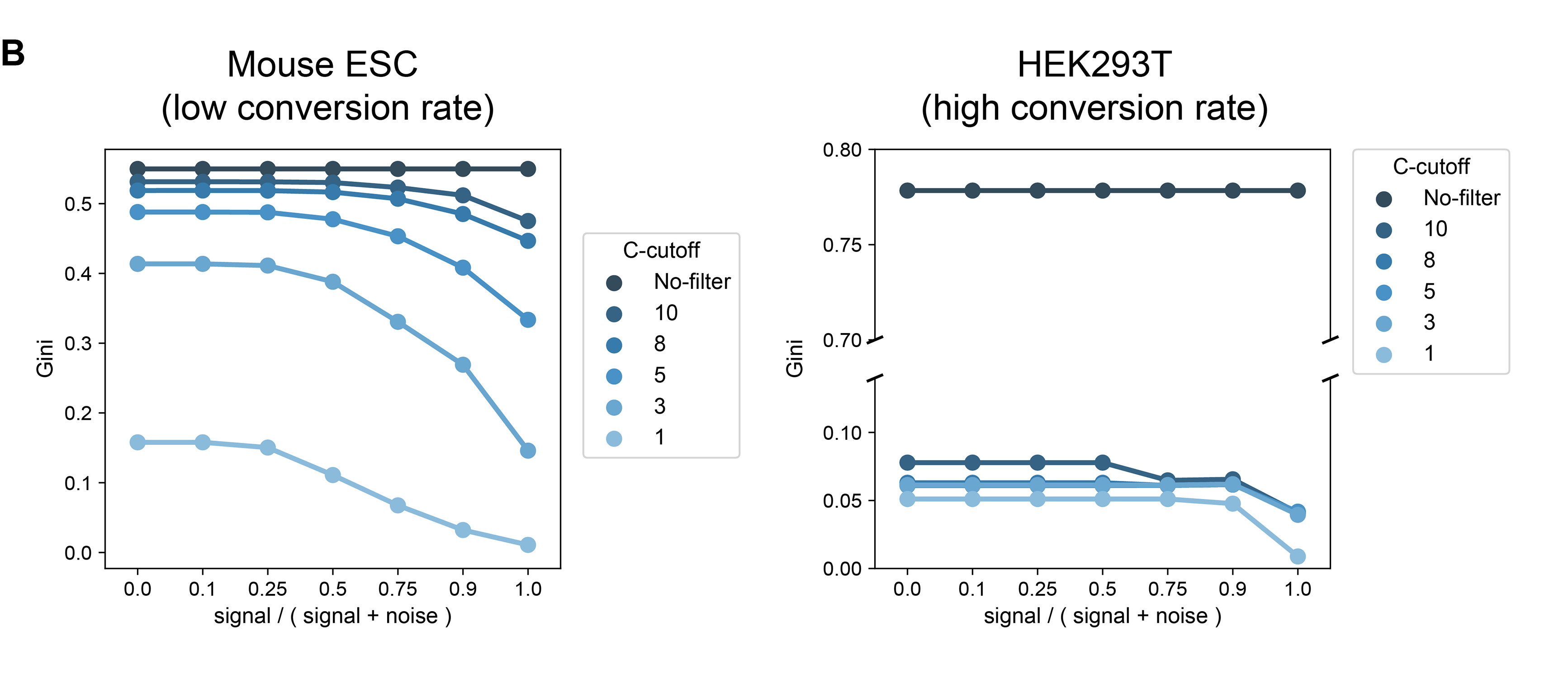
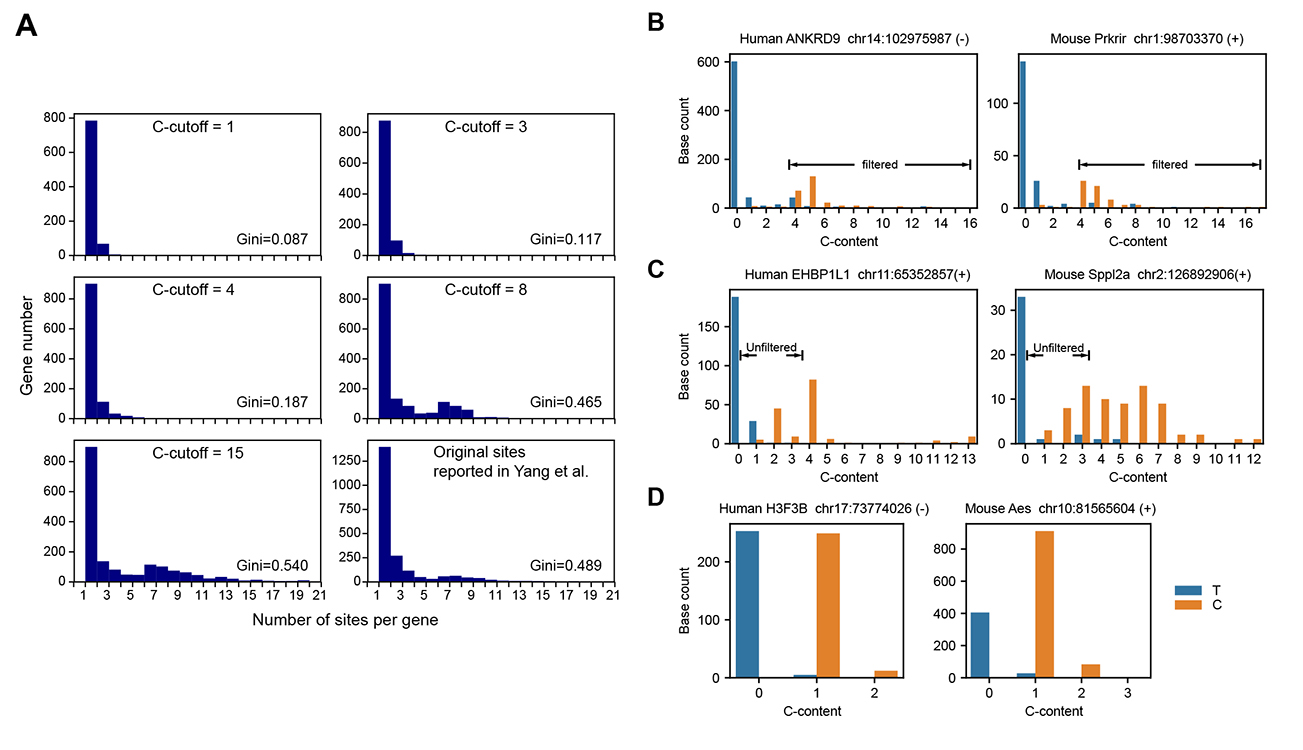
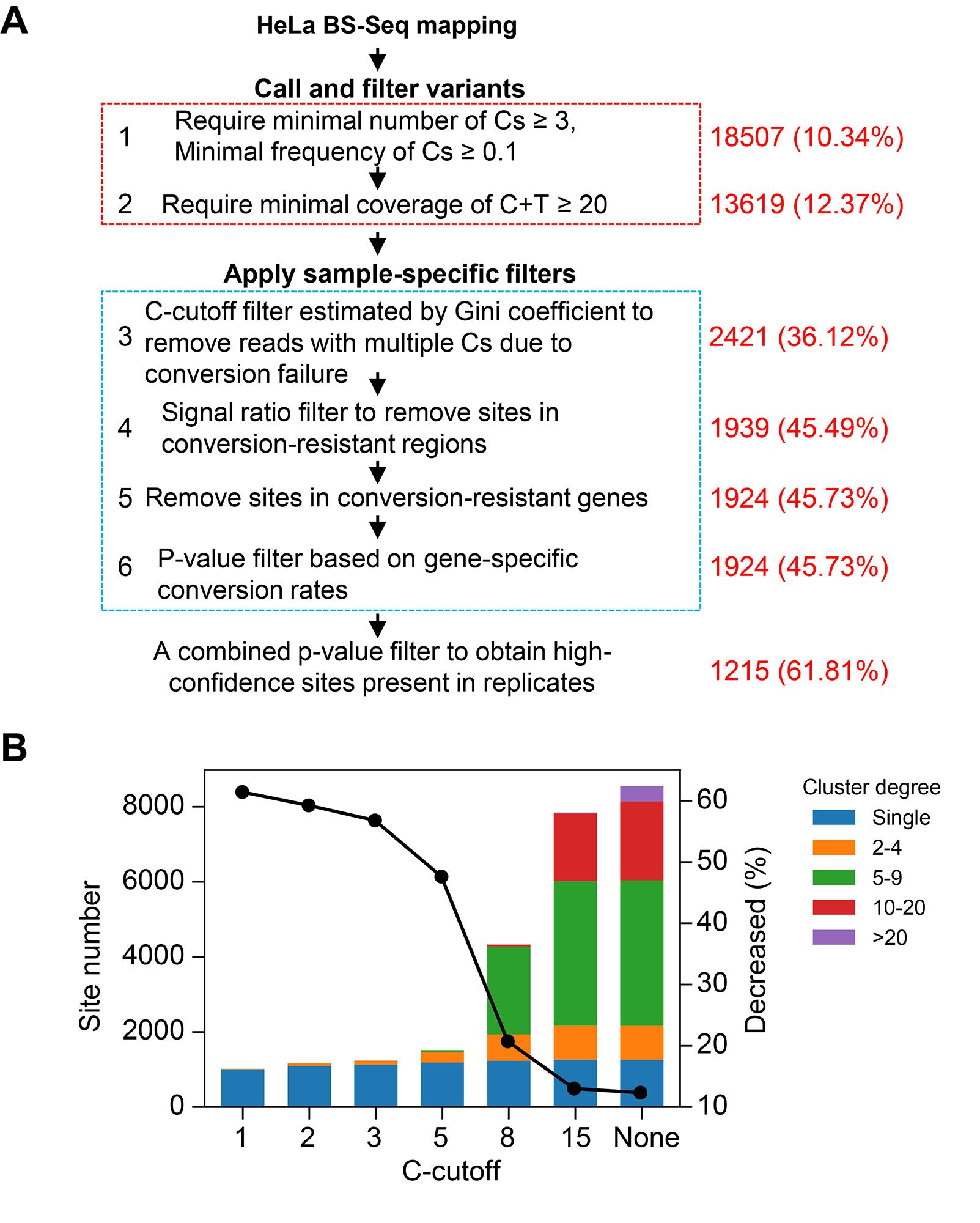
left, why gene-specific conversion rates are important; middle, gini-index vs signal rates in badly converted samples and well converted samples. And some examples for true positives and false positives; right, our filters can work perfectly!