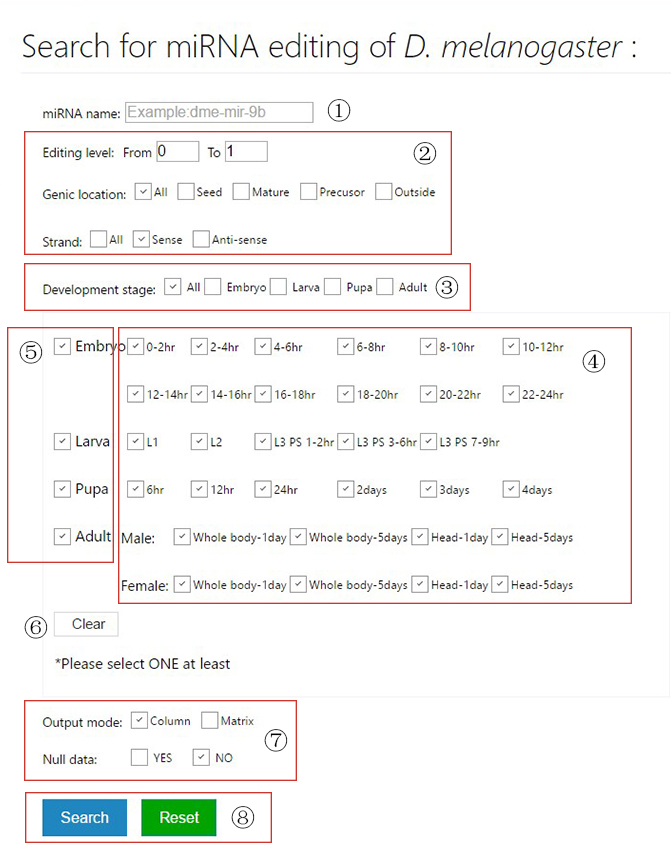
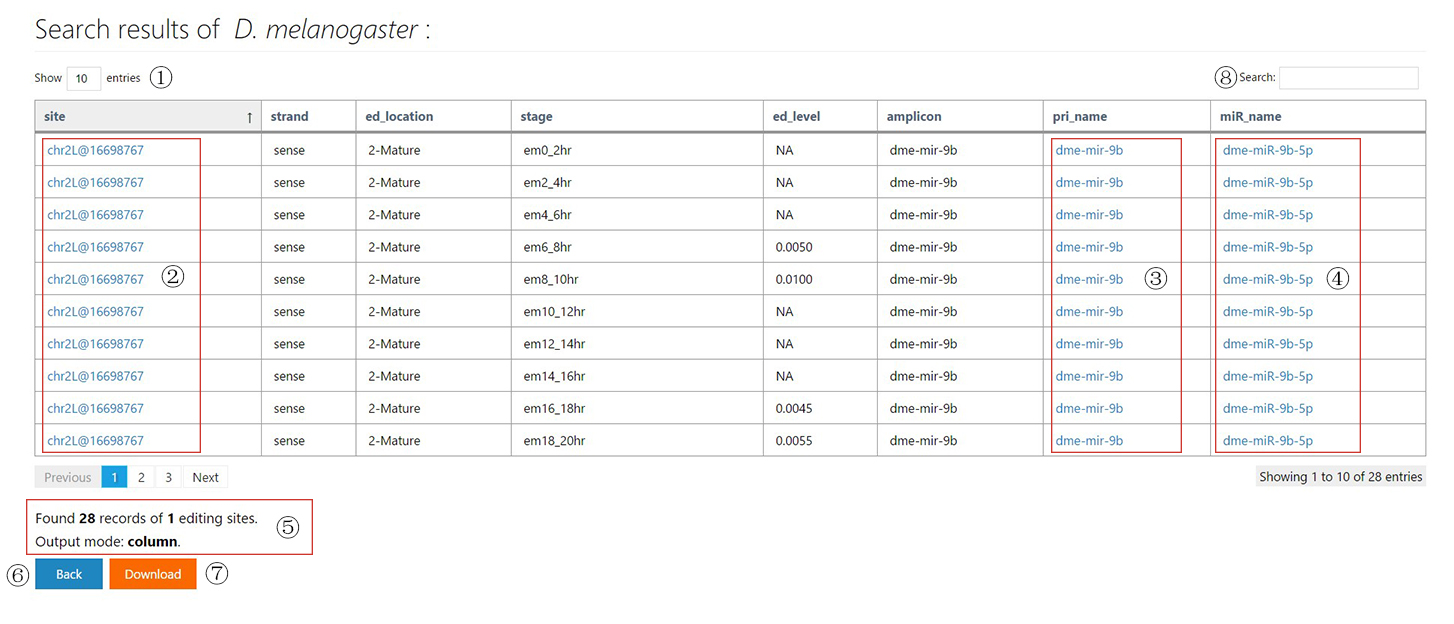
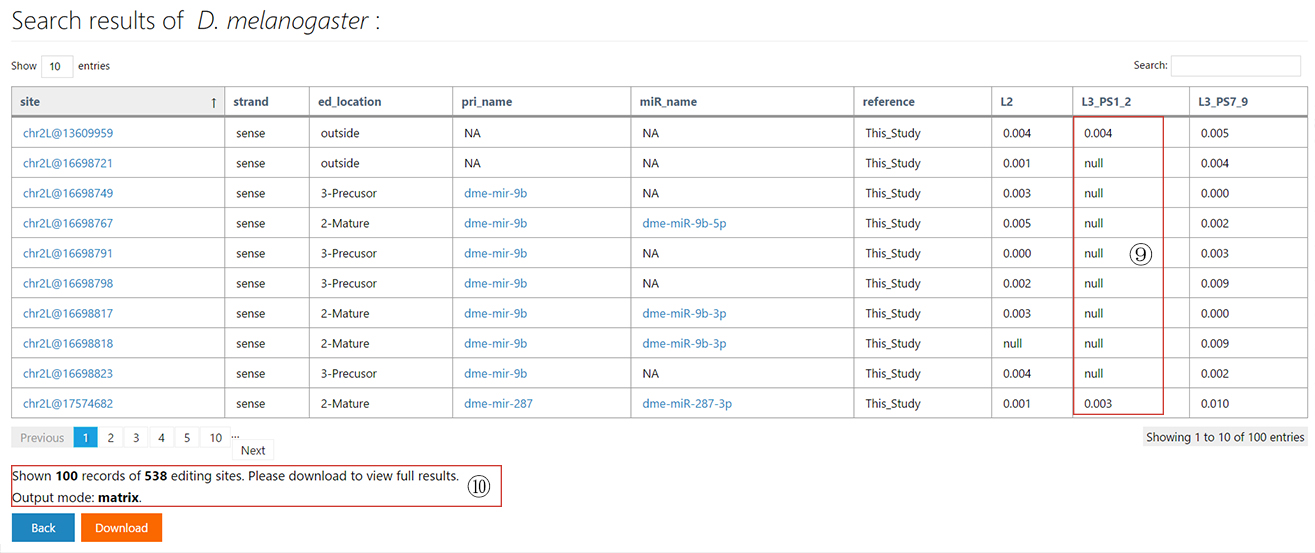
The miRNA editing database
The miRNA editing database (miREDB) was built with Asp, deployed on Windows Sever.
In this project, I learned a lot about HTML, SQL, Java script, CSS, and most importantly how to consider the project from the view of user , and especially how to prevent the user from destorying your sever. To be honest, the database was a bit slow in processing the inquires, because the codes writen were too heavy, but it indeed can run. No long after our paper published, we were forced to terminate our service as well as the lab home page deployed on the server, for the unlimited requirement of security from the bureaucrats.
The tutorial page and example search results of miREDB.
And the homepage of Zhanglab.
References
2018
-
The landscape of miRNA editing in animals and its impact on miRNA biogenesis and targeting
Lishi Li , Yulong Song , Xinrui Shi , and 7 more authors
Genome research, 2018
Adenosine-to-inosine (A-to-I) RNA editing regulates miRNA biogenesis and function. To date, fewer than 160 miRNA editing sites have been identified. Here, we present a quantitative atlas of miRNA A-to-I editing through the profiling of 201 pri-miRNA samples and 4694 mature miRNA samples in human, mouse, and Drosophila. We identified 4162 sites present in 80% of the pri-miRNAs and 574 sites in mature miRNAs. miRNA editing is prevalent in many tissue types in human. However, high-level editing is mostly found in neuronal tissues in mouse and Drosophila. Interestingly, the edited miRNAs in neuronal and non-neuronal tissues in human gain two distinct sets of new targets, which are significantly associated with cognitive and organ developmental functions, respectively. Furthermore, we reveal that miRNA editing profoundly affects asymmetric strand selection. Altogether, these data provide insight into the impact of RNA editing on miRNA biology and suggest that miRNA editing has recently gained non-neuronal functions in human.
Back