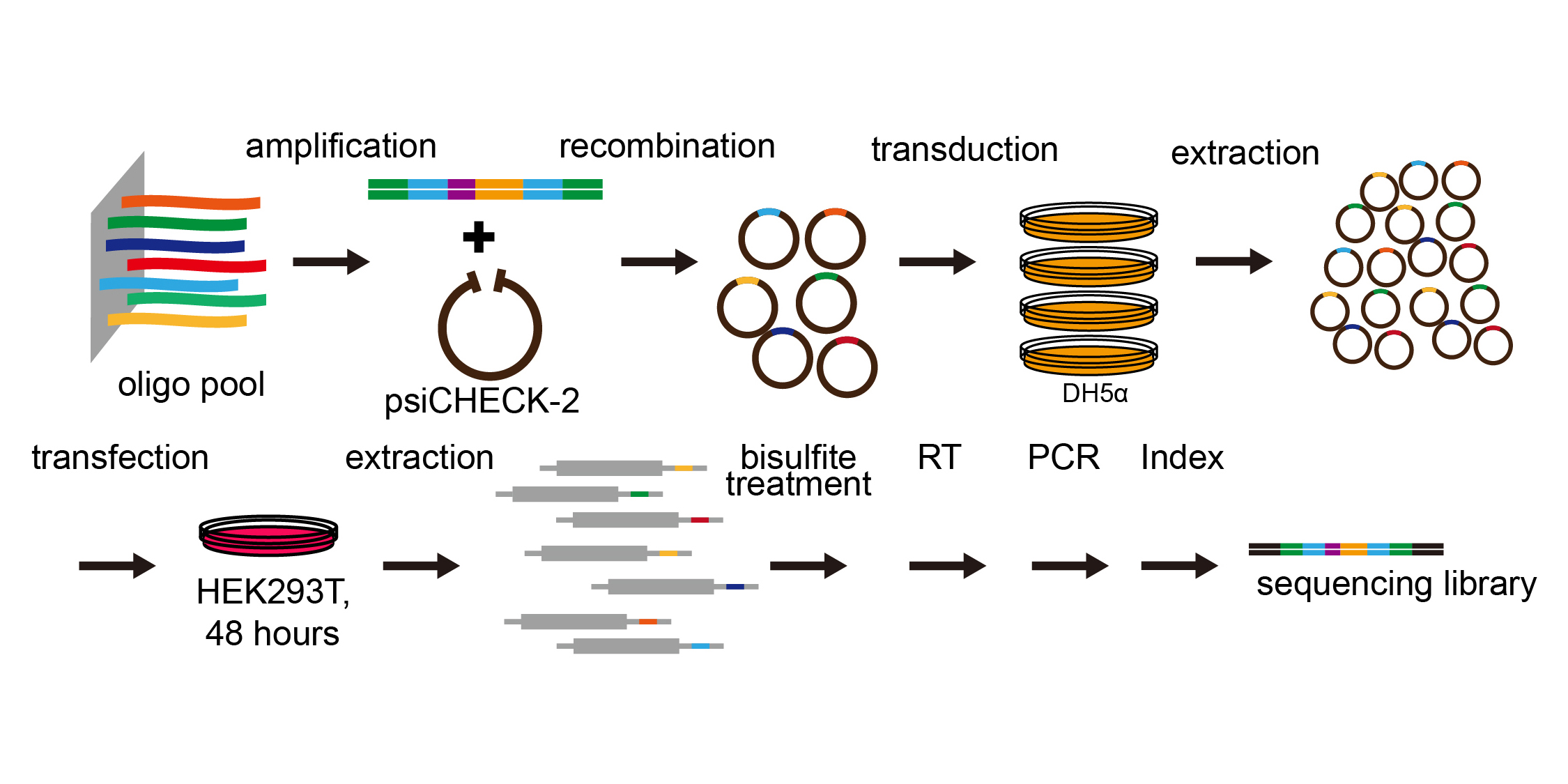
The workflow of high-throughput mutation assay: we synthesized the oligos, inserted them into a vector, purify the library, transfect them into the cells, and get reporter methylation levels from sequencing.
This is not a new strategy. In a previous paper studying pseudouridine, they adopt a same strategy to draw the consensus modification substrate of TRUB1. Here, we used pools to analyze the substrate recognition patterns of NSUN2 and NSUN6. A brief summary:
- NSUN2
- NSUN2 requires a stable stem loop in the 3’ downstream adjacent to the m5C sites.
- More Gs in the stem loop will result in higher methylation levels, but G is not so necessary for methylation in some cases.
- NSUN6
- NSUN6 is very sensitive to the change in the core motif (Nm5CUCCA).
- NSUN6 can tolerate some +3 C-to-U changes.
- The mechanism of preferences in NSUN6-mRNA recognition can be explained by our structure model.
References
2022
-
[accpeted] Developmental mRNA m5C landscape and regulatory innovations of massive m5C modification of maternal mRNAs in animals
Jianheng Liu , Tao Huang , Wanying Chen , and 13 more authors
2022
m5C is one of the longest-known RNA modifications, however, its developmental dynamics, functions, and evolution in mRNAs remain largely unknown. Here, we generate quantitative mRNA m5C maps at different stages of development in 6 vertebrate and invertebrate species and find convergent and unexpected massive methylation of maternal mRNAs mediated by NSUN2 and NSUN6. Using Drosophila as a model, we reveal that embryos lacking maternal mRNA m5C undergo cell cycle delays and fail to timely initiate maternal-to-zygotic transition, implying the functional importance of maternal mRNA m5C. From invertebrates to the lineage leading to humans, two waves of m5C regulatory innovations are observed: higher animals gain cis-directed NSUN2-mediated m5C sites at the 5’ end of the mRNAs, accompanied by the emergence of more structured 5’UTR regions; humans gain thousands of trans-directed NSUN6-mediated m5C sites enriched in genes regulating the mitotic cell cycle. Collectively, our studies highlight the existence and regulatory innovations of a mechanism of early embryonic development and provide key resources for elucidating the role of mRNA m5C in biology and disease.
2021
-
Sequence-and structure-selective mRNA m5C methylation by NSUN6 in animals
Jianheng Liu , Tao Huang , Yusen Zhang , and 4 more authors
National Science Review, 2021
mRNA m5C, which has recently been implicated in the regulation of mRNA mobility, metabolism and translation, plays important regulatory roles in various biological events. Two types of m5C sites are found in mRNAs. Type I m5C sites, which contain a downstream G-rich triplet motif and are computationally predicted to be located at the 5’ end of putative hairpin structures, are methylated by NSUN2. Type II m5C sites contain a downstream UCCA motif and are computationally predicted to be located in the loops of putative hairpin structures. However, their biogenesis remains unknown. Here we identified NSUN6, a methyltransferase that is known tomethylate C72 of tRNAThr and tRNA=Cys, as an mRNAmethyltransferase that targets Type II m5C sites. Combining the RNA secondary structure prediction, miCLIP, and results from a high-throughput mutagenesis analysis, we determined the RNA sequence and structural features governing the specificity of NSUN6-mediated mRNA methylation. Integrating these features into an NSUN6-RNA structural model, we identified an NSUN6 variant that largely loses tRNA methylation but retains mRNA methylation ability. Finally, we revealed a weak negative correlation between m5C methylation and translation efficiency. Our findings uncover that mRNA m5C is tightly controlled by an elaborate two-enzyme system, and the protein-RNA structure analysis strategy established may be applied to other RNA modification writers to distinguish the functions of different RNA substrates of a writer protein.
Back