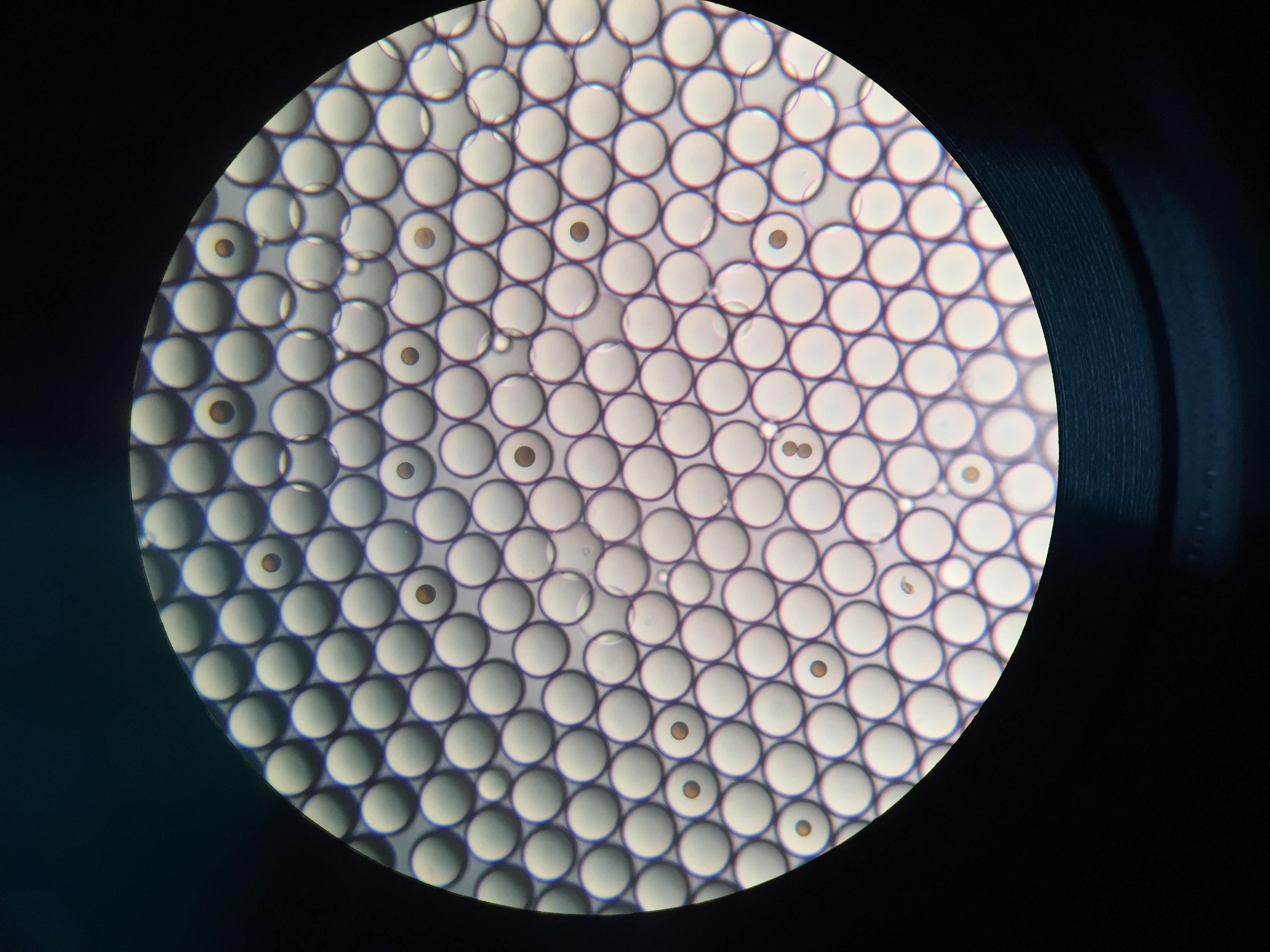
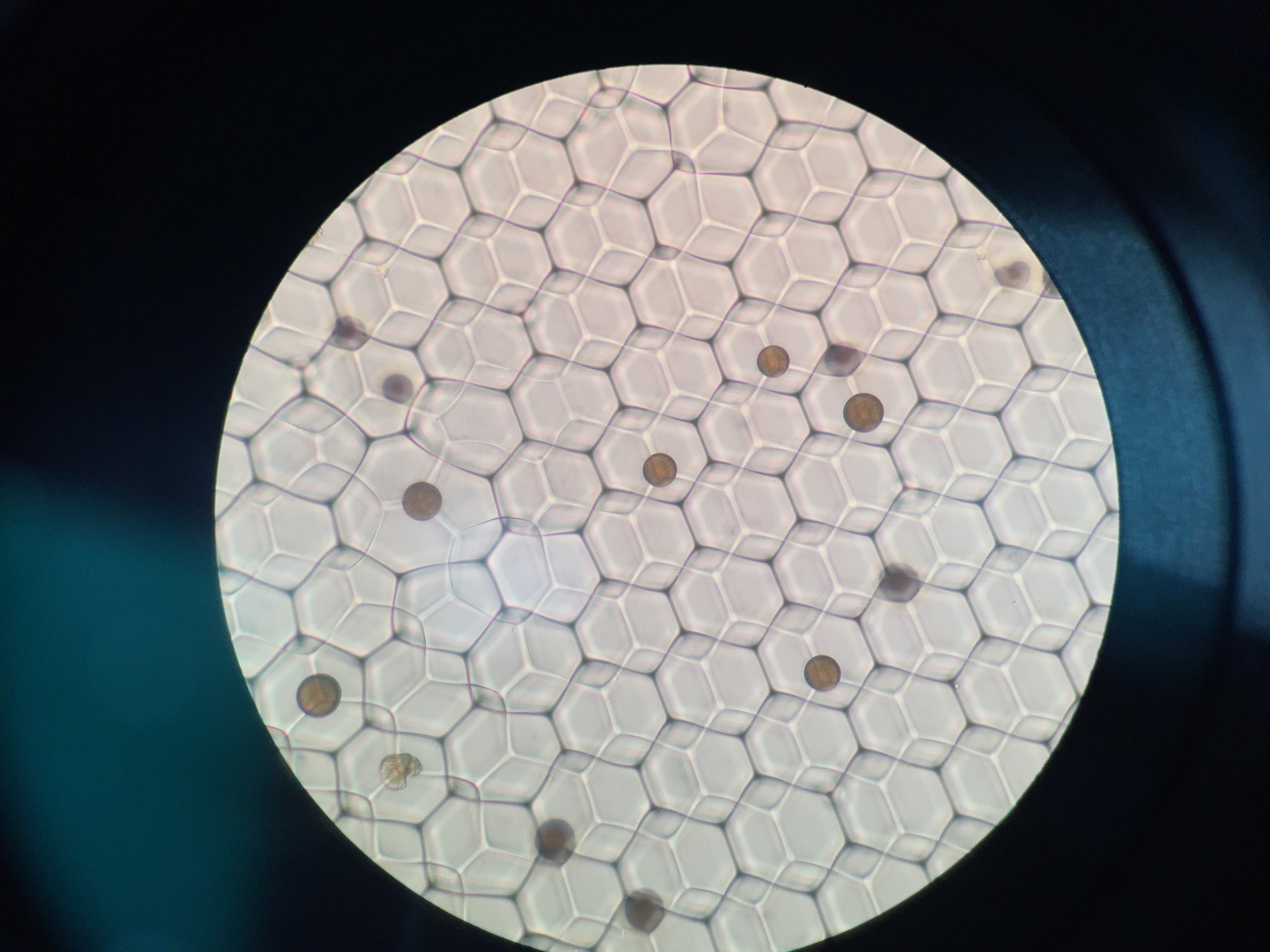
Drop-seq
Undergraduate thesis
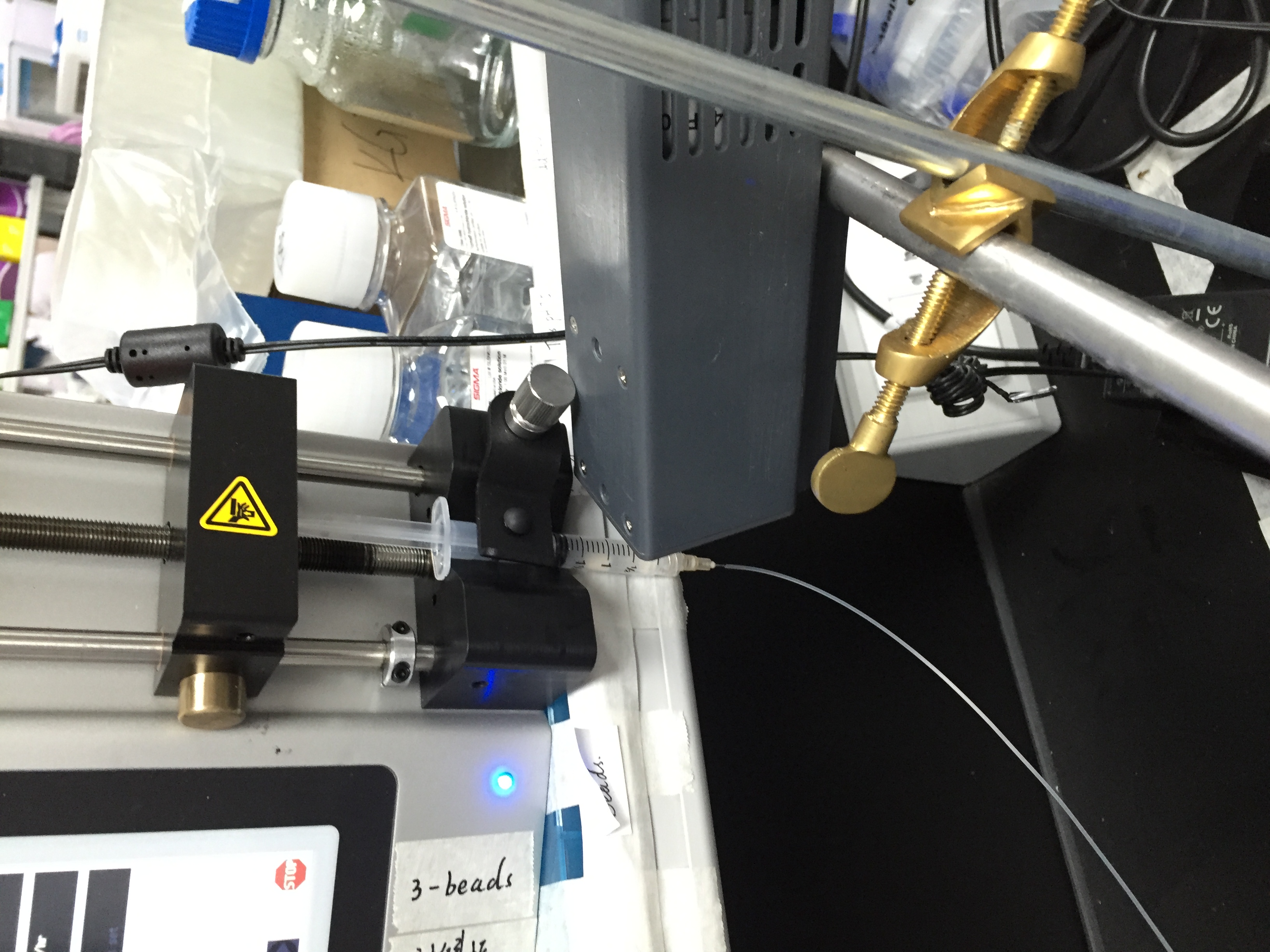
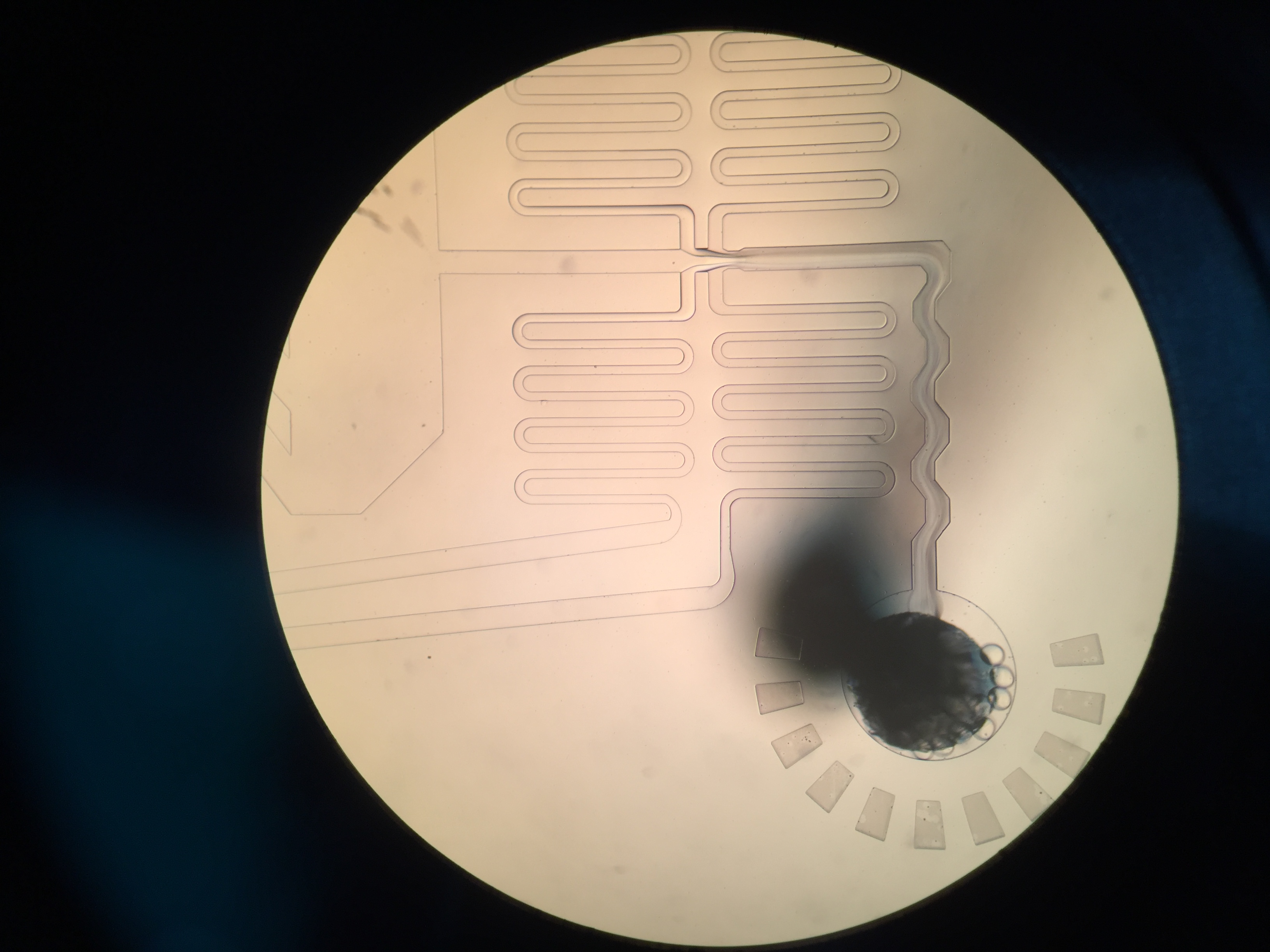
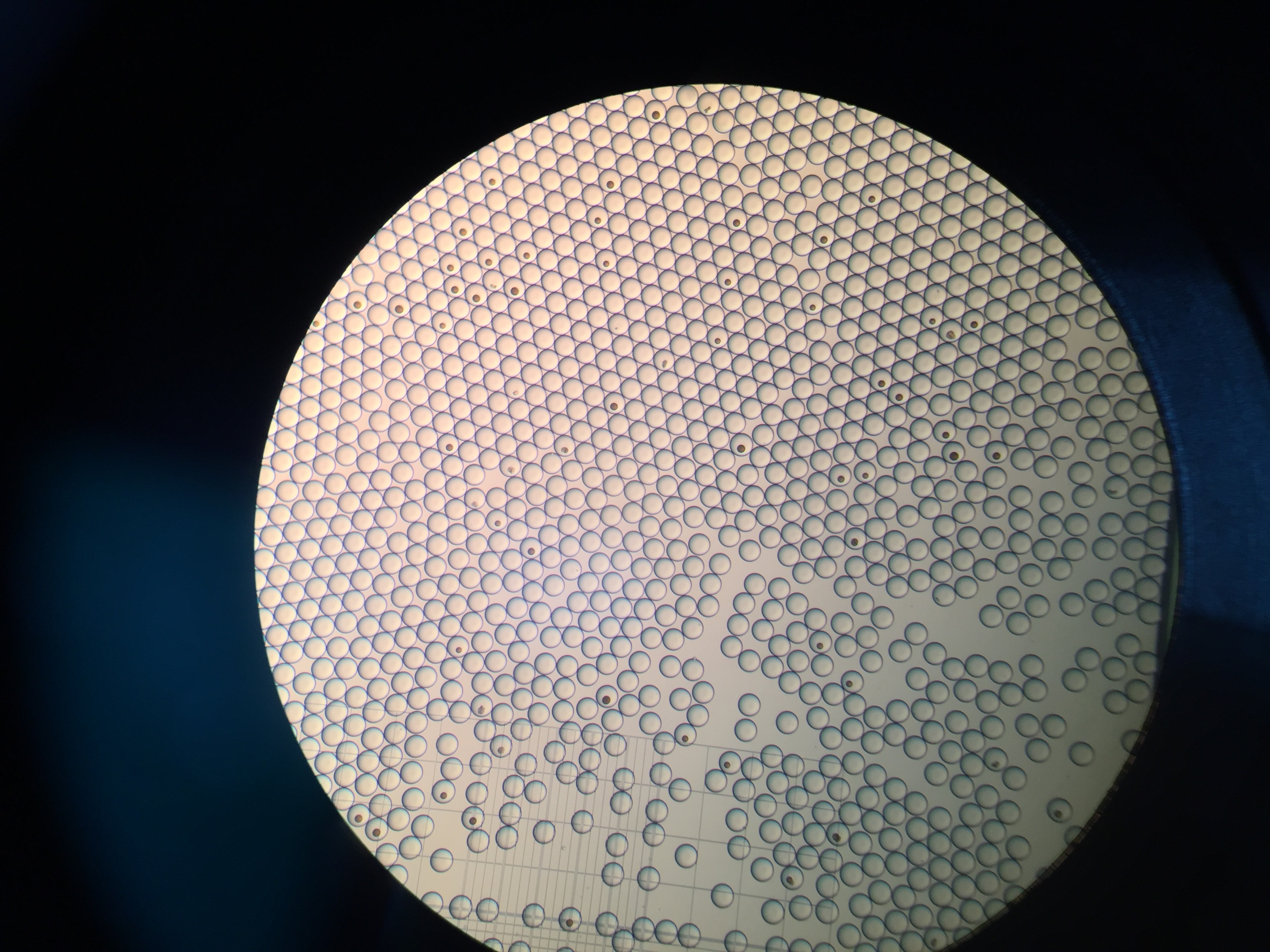
Drop-seq is my first experimental project at Zhang lab. It’s a happy experience, although we got nothing at the end. I learn a lot about DIY in biology. I try to do everything based on the guidelines, including contacting the sales all around the world. I guess nobody knows small RNA ligation more than me :).
We want to modify drop-seq for miRNA capture, but it’s a bit hard because the cost is too high (ligases) and the buffer is not compatible in practices (Sarkosyl will crystalize in the existence of Mg2+). But anyways, thanks Macosko for all the things (protocols, googel forum, etc.). About 1 year later, another group try to capture miRNA at single cell level via FACS sorting, and it seems no other groups interested in this these years.
Yes, we have some biological questions. What we want to know is about the distribution of RNA editing at single cell level: if RNA editing happens in a linear distribution among cells, or they distribute binarized? And this question has been answered via smFISH and other methods now.